DICOM - Fixing a bad DBSWin conversion.
posted by Neil Wallace - 1 Dec 2017, 11:45 a.m.
On the 19th January 2017, the teckies at Software of Excellence dialled into my former practice and converted the DBSWin image data over to a dicom format.
It was not done very well, but the dicom format gives interesting possibilities.
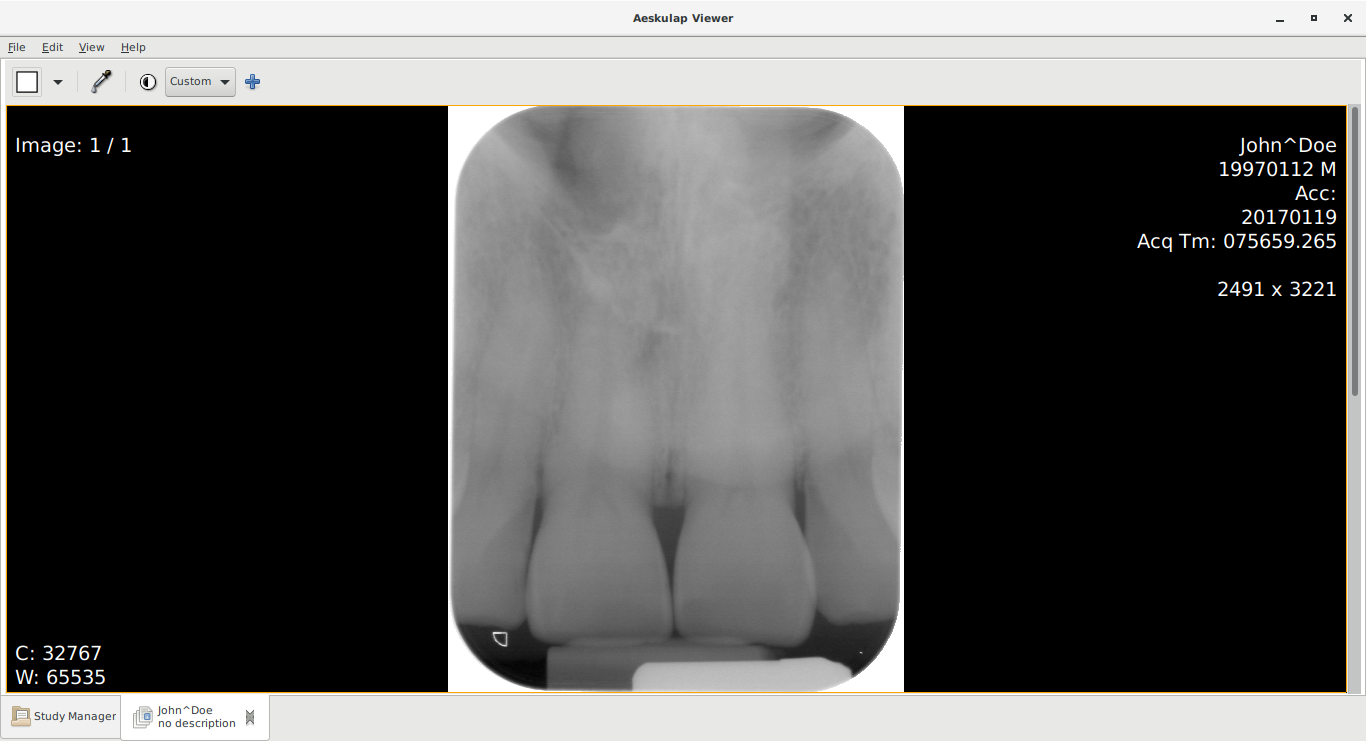
Image metadata in the *.dic files labelled every patient as
- NAME
- John Doe
- ID
- 123-45-6789
- SEX
- MALE
- DOB
- 12th January 1997
- DATE TAKEN
- 19th January 2017
This resulted in confusion (potentially litigation!) when viewing radiographs.
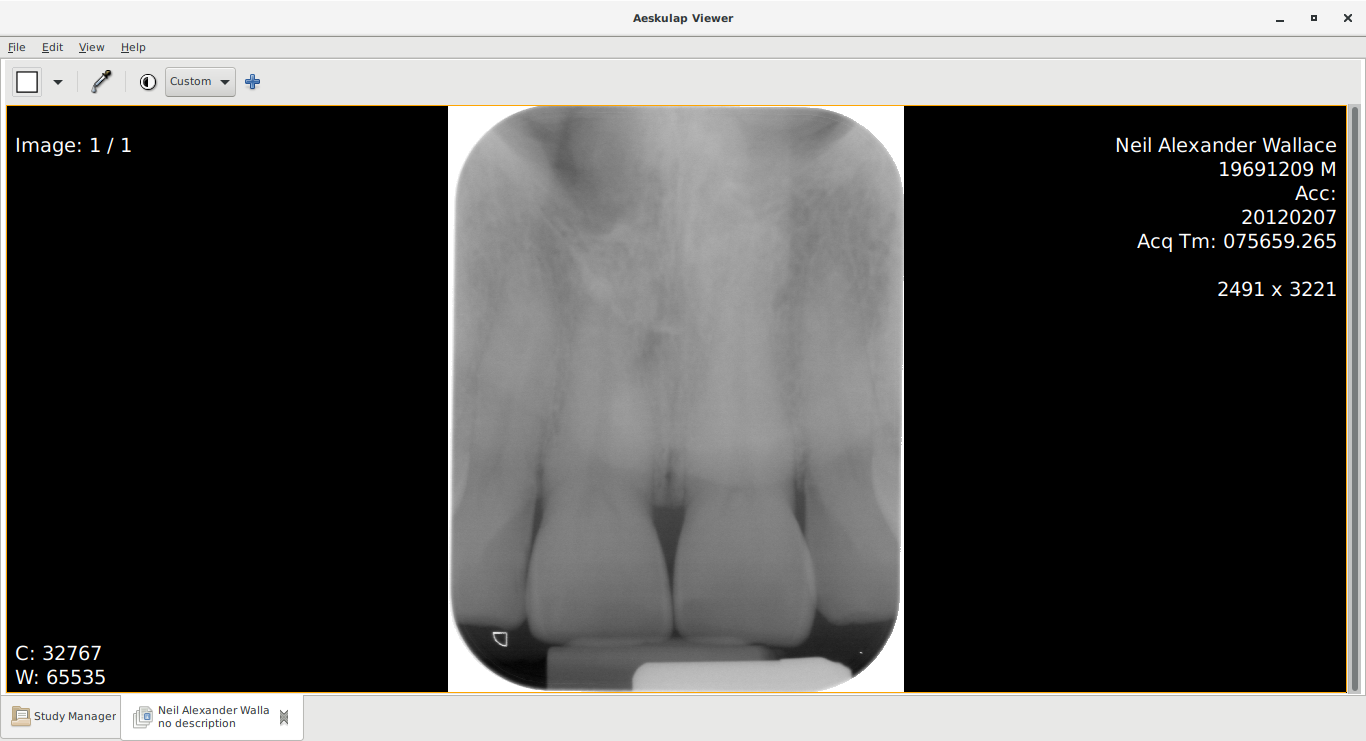
Below you can see the "before" and "after" view.
Before - who is this "John Doe"??
After - metadata updated.
 (Thanks to the pydicom module - I can access this trivially - along with a raft of extra parameters)
(Thanks to the pydicom module - I can access this trivially - along with a raft of extra parameters)
>>>import dicom
>>>dicom.read_file("11956/4094643865621.dic")
(0008, 0000) Group Length UL: 198
(0008, 0016) SOP Class UID UI: Secondary Capture Image Storage
(0008, 0018) SOP Instance UID UI: 1.2.840.114257.1.19012017.75659.1.1.1
(0008, 0020) Study Date DA: '20170119'
(0008, 0030) Study Time TM: '075659.265'
(0008, 0050) Accession Number SH: '1'
(0008, 0060) Modality CS: 'OT'
(0008, 0064) Conversion Type CS: 'DV'
(0008, 0070) Manufacturer LO: 'LEAD Technologies, Inc.'
(0008, 0090) Referring Physician's Name PN: 'Reffering^Doe'
(0010, 0000) Group Length UL: 62
(0010, 0010) Patient's Name PN: 'John^Doe'
(0010, 0020) Patient ID LO: '123-45-6789'
(0010, 0030) Patient's Birth Date DA: '19970112'
(0010, 0040) Patient's Sex CS: 'M'
(0020, 0000) Group Length UL: 116
(0020, 000d) Study Instance UID UI: 1.2.840.114257.1.19012017.75659.1
(0020, 000e) Series Instance UID UI: 1.2.840.114257.1.19012017.75659.1.1
(0020, 0010) Study ID SH: '1'
(0020, 0011) Series Number IS: '1'
(0020, 0013) Instance Number IS: '1'
(0028, 0000) Group Length UL: 196
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0008) Number of Frames IS: '1'
(0028, 0010) Rows US: 3221
(0028, 0011) Columns US: 2491
(0028, 0100) Bits Allocated US: 16
(0028, 0101) Bits Stored US: 16
(0028, 0102) High Bit US: 15
(0028, 0103) Pixel Representation US: 0
(0028, 0106) Smallest Image Pixel Value US: 0
(0028, 0107) Largest Image Pixel Value US: 65535
(0028, 1050) Window Center DS: '32767.'
(0028, 1051) Window Width DS: '65535.'
(0028, 1052) Rescale Intercept DS: '0.'
(0028, 1053) Rescale Slope DS: '1.'
(0028, 1054) Rescale Type LO: 'UNSPECIFIED'
(0028, 1055) Window Center & Width Explanation LO: ''
(7fe0, 0000) Group Length UL: 10411632
(7fe0, 0010) Pixel Data OB: Array of 10411612 bytes
In addition - all timestamps are set as the date when the conversion occurred, so the 1st job is to reset these so that directories can be perused sensibly.
neil@slim-silver /media/neil/HP_Pocket_Media_Drive/EXACTData/Xrays $ ls -lR 11956 11956: total 64M -rwxrwxrwx 2 neil neil 10M Jan 19 2017 4094643865621.dic -rwxrwxrwx 2 neil neil 605 Jan 19 2017 4094643865621.mih -rwxrwxrwx 2 neil neil 15M Jan 19 2017 '40946451537Bitewing-R(M).dic' -rwxrwxrwx 2 neil neil 658 Jan 19 2017 '40946451537Bitewing-R(M).mih' -rwxrwxrwx 2 neil neil 15M Jan 19 2017 '40946451985Bitewing-L(M).dic' -rwxrwxrwx 2 neil neil 658 Jan 19 2017 '40946451985Bitewing-L(M).mih' -rwxrwxrwx 2 neil neil 478K Jan 19 2017 4094646678727.jpg -rwxrwxrwx 2 neil neil 598 Jan 19 2017 4094646678727.mih -rwxrwxrwx 2 neil neil 447K Jan 19 2017 '41088486955Video undefiniert.jpg' -rwxrwxrwx 2 neil neil 673 Jan 19 2017 '41088486955Video undefiniert.mih' -rwxrwxrwx 2 neil neil 8.1M Feb 2 2017 '42768360239OPG undefiniert.dic' -rwxrwxrwx 2 neil neil 695 Feb 2 2017 '42768360239OPG undefiniert.mih' -rwxrwxrwx 2 neil neil 3.3M Feb 8 2017 42774602478RX01.DIC -rwxrwxrwx 2 neil neil 3.4M Feb 8 2017 42774602478RX01_M.DIC -rwxrwxrwx 2 neil neil 806 Feb 8 2017 42774602478RX01.mih -rwxrwxrwx 1 neil neil 321 Jul 13 14:24 Patient.inf
After running this script this problem is solved.
neil@slim-silver /media/neil/HP_Pocket_Media_Drive/EXACTData/Xrays $ ls -lR 11956 11956: total 64M -rwxrwxrwx 2 neil neil 10M Feb 7 2012 4094643865621.dic -rwxrwxrwx 2 neil neil 605 Feb 7 2012 4094643865621.mih -rwxrwxrwx 2 neil neil 15M Feb 7 2012 '40946451537Bitewing-R(M).dic' -rwxrwxrwx 2 neil neil 658 Feb 7 2012 '40946451537Bitewing-R(M).mih' -rwxrwxrwx 2 neil neil 15M Feb 7 2012 '40946451985Bitewing-L(M).dic' -rwxrwxrwx 2 neil neil 658 Feb 7 2012 '40946451985Bitewing-L(M).mih' -rwxrwxrwx 2 neil neil 478K Feb 7 2012 4094646678727.jpg -rwxrwxrwx 2 neil neil 598 Feb 7 2012 4094646678727.mih -rwxrwxrwx 2 neil neil 447K Jun 28 2012 '41088486955Video undefiniert.jpg' -rwxrwxrwx 2 neil neil 673 Jun 28 2012 '41088486955Video undefiniert.mih' -rwxrwxrwx 2 neil neil 8.1M Feb 2 2017 '42768360239OPG undefiniert.dic' -rwxrwxrwx 2 neil neil 695 Feb 2 2017 '42768360239OPG undefiniert.mih' -rwxrwxrwx 2 neil neil 3.3M Feb 8 2017 42774602478RX01.DIC -rwxrwxrwx 2 neil neil 3.4M Feb 8 2017 42774602478RX01_M.DIC -rwxrwxrwx 2 neil neil 806 Feb 8 2017 42774602478RX01.mih -rwxrwxrwx 1 neil neil 321 Jul 13 14:24 Patient.inf
Now that is done, I can use the dicom module to populate the patient name, date of birth, sex and ID fields trivially using the data from the .mih files and Patient.inf file.
I am planning that the next version of OpenMolar will be able to display these images.
